南科大医学院
官方公众号

返回首页:news center
更新时间:2020-04-27
New findings by researchers at Southern University of Science and Technology (SUSTech) could lead to a dramatic shift in the quality of medical care across the world. The use of digital metabolic mapping through quantitative systems biology provides the opportunity for scientists to understand every reaction inside the human body, and what it means for you.
On March 18, Research Assistant Professor Hemi Luan and Professor Wenyong Zhang (School of Medicine) worked with collaborators across the Greater Bay Area to publish a paper in the high-impact academic journal, Bioinformatics (IF =4.531). The article was titled “CPVA: a web-based metabolomic tool for chromatographic peak visualization and annotation.”
Quantitative systems biology (QSB) describes cells, tissues, organs, and various molecules with different structures and functions. With the human body being a highly complex network of metabolic and biochemical reactions, many different factors could influence the physiological state of the body, as well as disease function.
QSB takes a digital approach to examining the different molecules in the metabolic network nodes in constructing a computational model. A digital map would chart the life cycle of each molecule, tracking the metabolic network composed of genes and small molecular metabolites that are key for QSB. These small metabolites can more accurately reflect the instantaneous life cycle of the system.
The continuous improvement of omics methodology based on liquid chromatography-mass spectrometry (LC-MS) has resulted in the improvement of our understanding of metabolic networks. Rapid developments in this field have allowed scholars to analyze thousands of metabolites at once, but the current data collection system can result in many false positives.
There is an enormous need for intelligent processing of omics through algorithms and tools for the proper characterization of metabolites. The paper aims to minimize a large number of false positives and redundant signal peaks by proposing a chromatographic center strategy algorithm (CSS) while developing interactive tools, known as the CVPA.
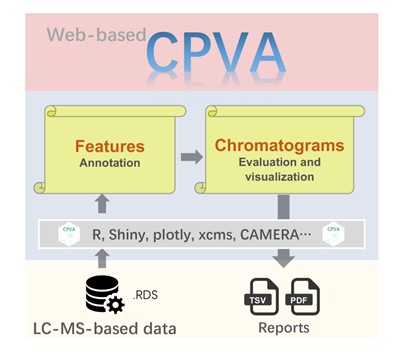
Figure 1: Flow chart of CPVA algorithm and online tools
The CCS algorithm assesses a series of issues, including symmetry, jaggedness, modality, and chromatography quality index (MCQ). Their method displays the morphological features of the peaks. The CVPA automatically tags stable isotope clusters, adducts, and common mass spectrometric background contaminants. Their design significantly reduces the number of false positives and redundant peaks while improving non-targeted metabolomics data.
The research team used the receiver operating curve (ROC) to evaluate the CVPA and its ability to process real data. Their results showed that the CVPA performed exceptionally well in identifying false positives or redundant peaks.
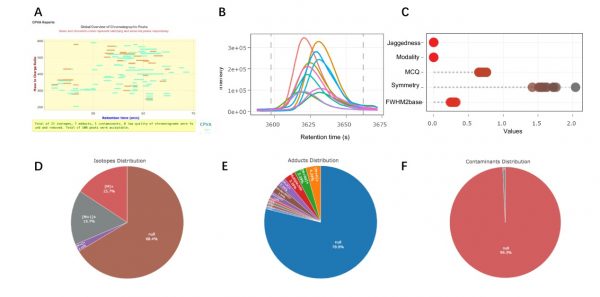
Figure 2: Results of CPVA algorithm and tools
SUSTech research assistant professor Hemi Luan was the first author. Professor Wenyong Zhang and Research Assistant Professor Hemi Luan were the correspondent authors. Additional contributors came from the Beijing University of Chinese Medicine Shenzhen Hospital, State Key Laboratory of Environmental and Biological Analysis at Hong Kong Baptist University (HKBU). The research received support from the National Natural Science Foundation of China and the Education Science Program of Shenzhen.
Algorithm and software links:
CPVA: http://cpva.eastus.cloudapp.azure.com
Source code and installation instructions are available on GitHub: https://github.com/13479776/cpva
Paper link: https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btaa200/5809525